Institution: ICB
Departement: DSCTM
Typology: Facility, Research field/topic, Skill, Software
Location: Via Campi Flegrei 34, 80078 Pozzuoli (NA), Italy
Contacts:
- Pietro Amodeo (pamodeo@icb.cnr.it)
- Rosa Maria Vitale (rmvitale@icb.cnr.it)
Description:
The main computational chemistry skills in the ICB Computational Chemistry and Cheminformatics group (ICB-CCC) cover the fields of homology modelling, molecular docking and molecular dynamics simulations of biomolecular systems and quantum chemistry calculations on isolated molecules or their complexes with representative protein fragments. These skills are mainly aimed at studying the driving forces at molecular level of ligand-protein interactions, the molecular bases of receptor selectivity, and the conformational transitions associated to the modulation of receptor activities. As for the main application areas of these techniques and approaches, ICB-CCC devotes a special attention to the characterization of nuclear receptors, GPCRs and ion-receptor channels in complex with different modulators, and to drug discovery projects. The latter are currently carried out by performing in silico screening of small libraries of compounds from both natural and synthetic source in order to: identify novel leads, determine structure-activity relationships and drive their optimization, in close synergy with organic chemists and pharmacologists. In the field of cheminformatics, ICB-CCC has developed and deployed the StOrMoDB platform, an integrated database and toolset to manage full lab workflows, including collection/production, extraction, purification, compound identification and activity assaying, of both natural products and molecular libraries from design/synthesis projects. The "Extensive H-Bond Analysis Software" (EHBAS) platform is a bioinformatic application currently under development at ICB-CCC to perform fully customizable batch processing of the whole Protein Data Bank (PDB) dataset aimed to search for H-bonds, including those involving solvent and other “small molecules,” and to provide reasonable assignment of possible interactions even for groups endowed with multiple conformations. Presently working as a set of standalone tools, EHBAS is under further development, to integrate the different components into a single program, to be deployed as a web service endowed with a user-friendly interface.
Team:
- Pietro Amodeo (pamodeo@icb.cnr.it)
- Rosa Maria Vitale (rmvitale@icb.cnr.it)
- Salvatore Donadio (salvatore.donadio@icb.cnr.it)
https://stormodb.na.icb.cnr.it/stormodb/
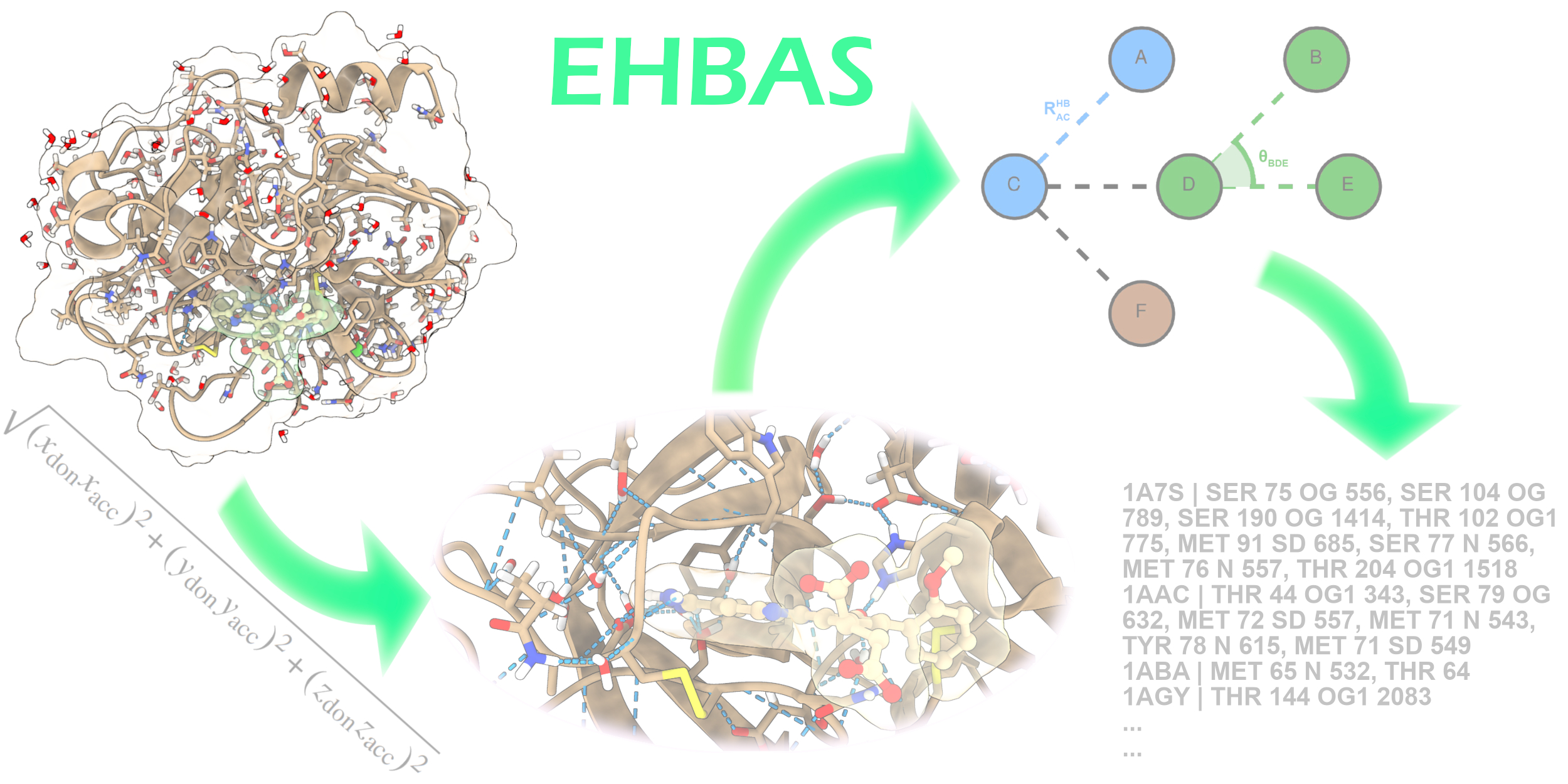
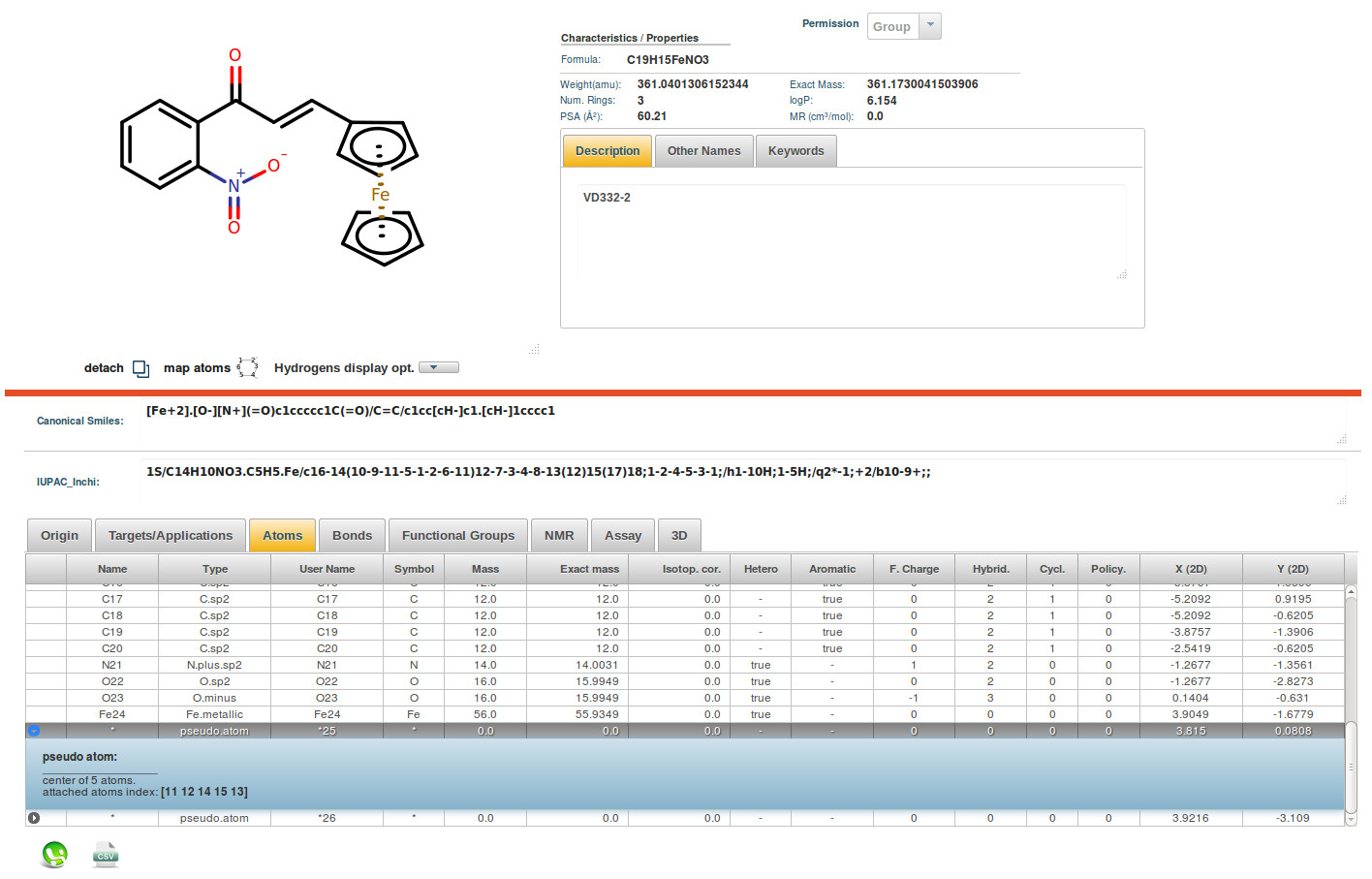

(TRPV1_MD_in_POPC_water_ions.png) Figure 1. A MD simulation snapshot of TRPV1 channel in complex with an agonist, performed in POPC membrane, water and ions environment. TRPV1 is shown as ribbons with stick bonds for residue sidechains involved in ligand binding and a fully-transparent protein surface with green outline. Agonist is depicted in stick representation. An overall transparent molecular surface is used for POPC (gold) and water (very light blue) molecules, while small solid spheres represent sodium (violet) and chlorine (green) atoms. (StOrMoDB_Molecule_page_example_Metal-organic_mol.png) Figure 2. Typical main page for molecular objects in StOrMoDB, shown for a metal-organic molecule (MOM). The platform can import and manage information about MOM's through an extension of the MDL mol v3000 format, featuring a full and consistent description of pseudo centers and coordination bonds. (EHBAS_workflow_Graphic_concept.png) Figure 3. Graphic concept for EHBAS workflow.
