Institution: IBF
Departement: DSFTM
Typology: Software
Location: Via de Marini 6, 16149 Genova
Contacts:
- Michael Pusch (michael.pusch@ibf.cnr.it)
- Alessandra Picollo (Alessandra.Picollo@ibf.cnr.it)
Description:
ALLIN - (Annotation of sequence aLignment and structuraL ProteIn visualizatioN) is a web interface to generate an interactive HTML web page for the simultaneously visualization of annotated protein sequences alignment and 3D structures or homology models of the related proteins. The physiological, functional, and structural properties of proteins and their pathogenic variants can be summarized using many tools. The amount of information relating to a single protein is large, often spread among different sources that require different programs to access. It is not always easy to select, simultaneously visualize, and compare specific properties of different proteins. On the other hand, comparing members of the same protein family could suggest conserved properties or highlight significant differences. We have thus developed a web interface, called ALLIN (Annotation of Sequence ALignment and structuraL ProteIn visualizatioN) to generate a web page for the simultaneous visualization of multi-sequence protein alignments, including comments and annotations, and the related three-dimensional protein structures. The distinctive feature of this interface is that it permits the inclusion of comments and coloring of residues in the alignment section, in accordance with a user-defined color code, allowing a quick overview of specific properties. The interface does not require training or coding expertise. The final result is a unique “memo” web page that combines data from different sources, with the flexibility to highlight only the information of interest. The output can provide a general overview of the state of art of a protein family that is easily shared among researchers with different backgrounds. Protein regions or single residues of interest can be quickly visualized to be analyzed in more detail using dedicated programs and new data can be conveniently added as it emerges. We believe the ALLIN tool can be useful for all scientists working on the structure-function analysis of proteins, in particular on those involved in human genetic diseases. The tool is described in the following publication: Picollo & Pusch, ALLIN: a tool for annotation of a protein alignment combined with structural visualization. 2024. J Gen Physiol, doi: 10.1085/jgp.202413635 Please cite the publication when you utilize ALLIN. This work was supported in part by the Italian Ministry of Foreign Affairs and International Cooperation”, grant number SE24GR07
Team:
- Michael Pusch
- Alessandra Picollo
https://github.com/mikpusch/ALLIN
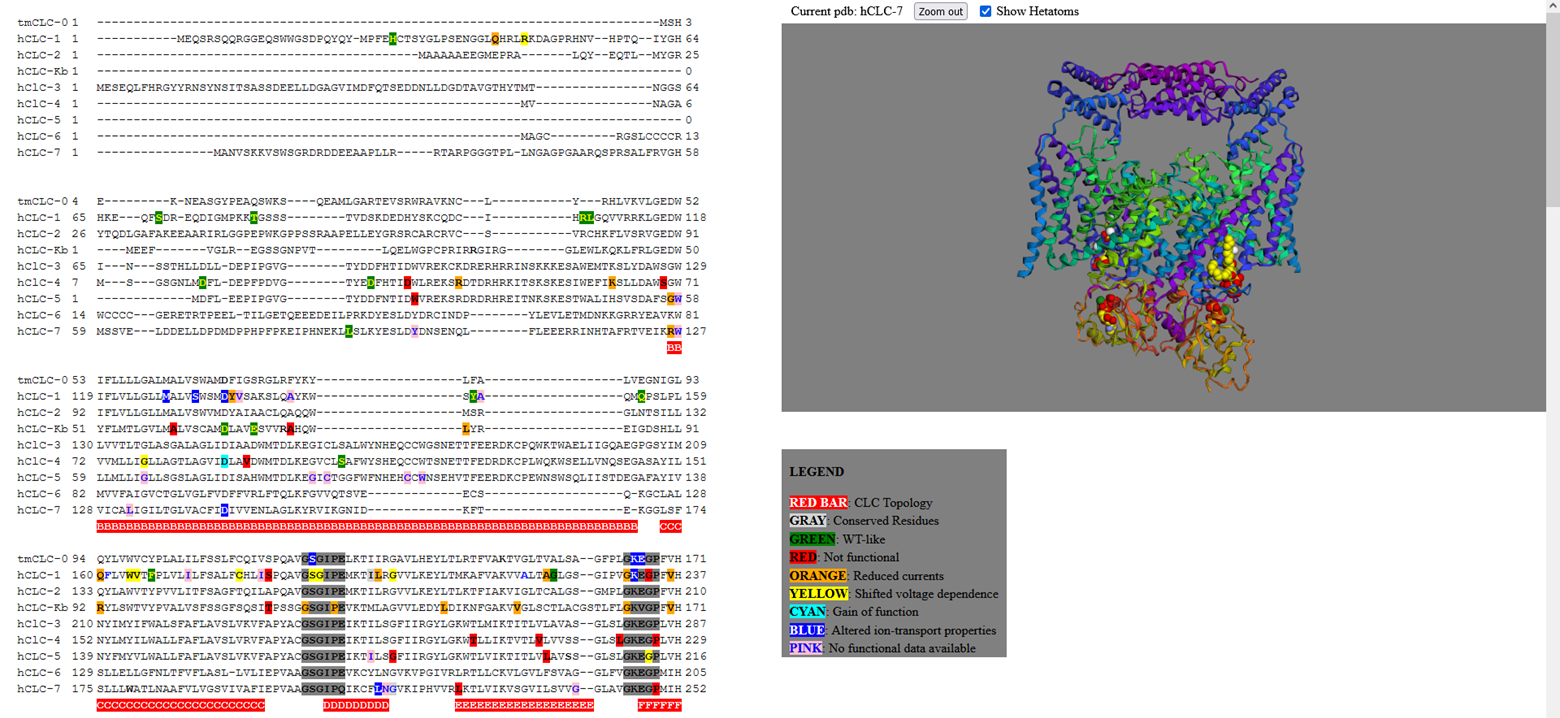
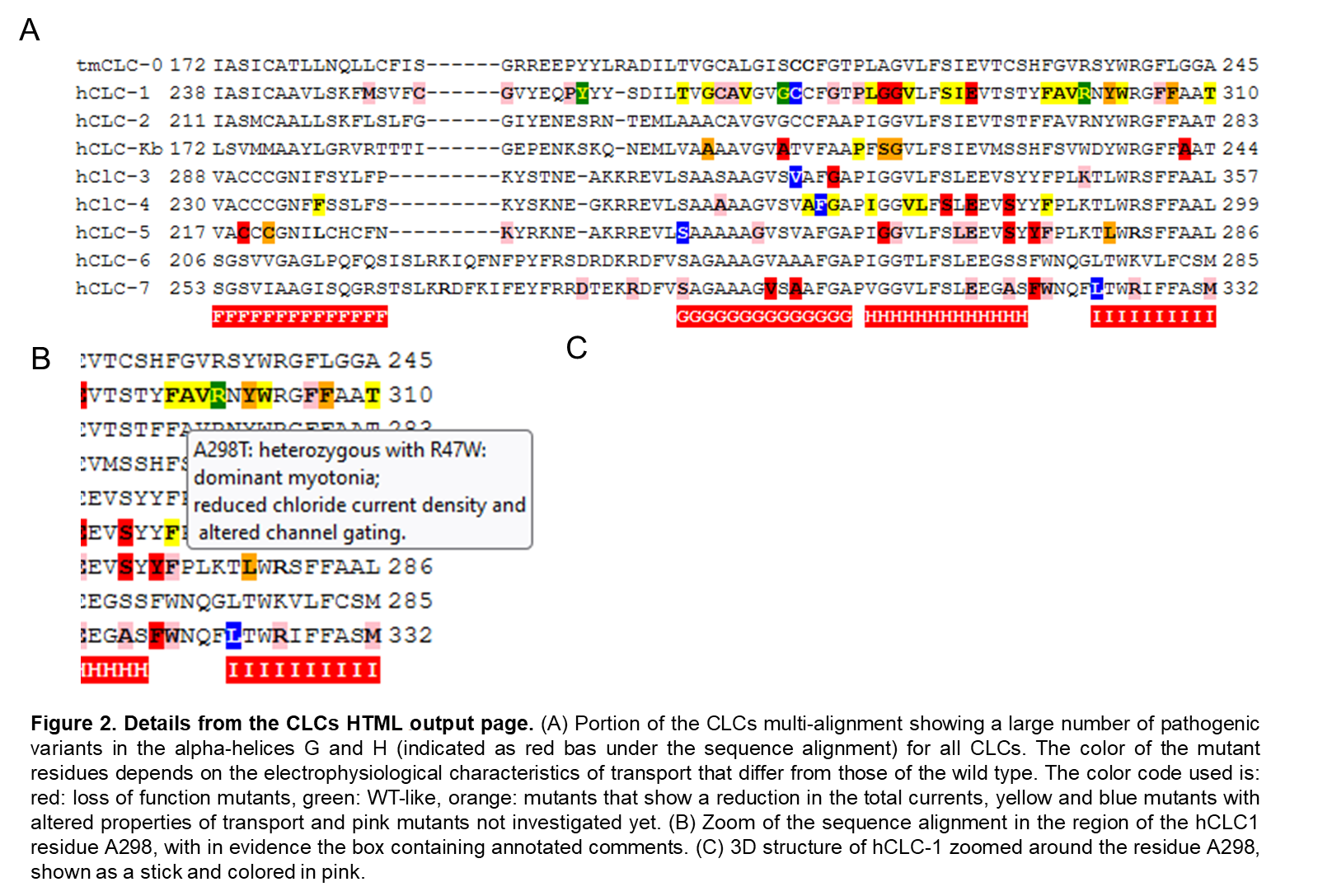
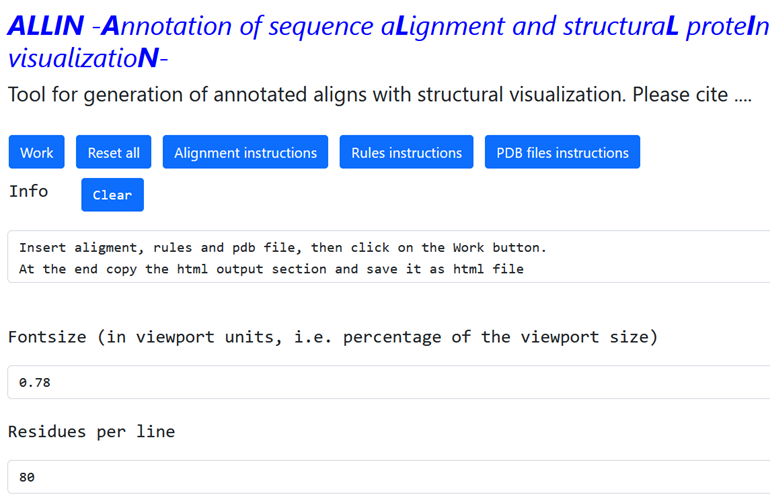
1 - Unique HTML page with annotated sequence multi-alignment and protein structure visualization. A screenshot of the browser windows generated for the CLC chloride transporting protein family is shown. On the left, a section of the multiple protein alignment is captured with the insertion of graphical annotations better explained in figure 2. On the right the legend relative to the colosr present in the sequence alignment. At the top of the page, it is indicated which pdb file is currently loaded to show the corresponding 3D protein structure in the molecular visualization box, with the included option to show or not Hetatoms 2-Details from the CLCs HTML output page. (A) Portion of the CLCs multi-alignment showing a large number of pathogenic variants in the alpha-helices G and H (indicated as red bas under the sequence alignment) for all CLCs. The color of the mutant residues depends on the electrophysiological characteristics of transport that differ from those of the wild type. The color code used is: red: loss of function mutants, green: WT-like, orange: mutants that show a reduction in the total currents, yellow and blue mutants with altered properties of transport and pink mutants not investigated yet. (B) Zoom of the sequence alignment in the region of the hCLC1 residue A298, with in evidence the box containing annotated comments. (C) 3D structure of hCLC-1 zoomed around the residue A298, shown as a stick and colored in pink. 3-Overview of the ALLIN web interface. View of the three sections of the web interface. (A) Section 1 contains the info box and the two boxes that define the fontsize and the number of residues per line that will be shown in the multi-sequence alignment. At the top five buttons: to start the procedure (“work”), to clean all (“Reset all”) and three instruction buttons, are present (B) The three input boxes with example of fasta alignment, string of rules and protein atomic coordinates. (C) Example of HTML output script.
